install.packages('did')did
R package for doubly-robust and ipw DiD estimator (callaway and sant’anna 2021)
did is a package that implements the DiD estimators (doubly-robust, inverse probability weighting) proposed by Callaway and Sant’Anna (2021) that solves the bias of the TWFE estimator in staggered DiD. Documentation can be found here.
Install the package as follows:
sample code
Start by loading packages and the data:
# packages needed:
library(did)
library(readr) # for importing data
df = read_csv('df.csv')We use the att_gt() function to run the matching process of csdid. We can use the doubly-robust (dr) method, or the inverse probability weighing (ipw) method by changing the argument of est_method =.
Notes: id variable should be transformed to an integer variable before starting. Set cohort = 0 for never-treated units. Generally, doubly-robust is the better estimator, so use doubly-robust unless there is an error.
mod = att_gt(
# required arguments
yname = "outcome",
tname = "time",
idname = "id", # must be a integer-variable
gname = "cohort", # cohort = 0 for never-treated
est_method = "dr", # change to ipw if you are having issues
base_period = "universal", # do not change
allow_unbalanced_panel = T, # generally good to keep this T
data = df,
xformla = ~ covar, # (optional)
control_group = "nevertreated", # use "notyettreated" if sample size is small
panel = T # change to F if you are using rep. cross-section
)We use the aggte() function to aggregate our matched treatment effects into an overall treatment effect.
mod |>
aggte(type = "simple", na.rm = T) |>
summary(att)#>
#> Call:
#> aggte(MP = mod, type = "simple", na.rm = T)
#>
#> Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
#>
#>
#> ATT Std. Error [ 95% Conf. Int.]
#> -1.1237 0.4887 -2.0815 -0.1659 *
#>
#>
#> ---
#> Signif. codes: `*' confidence band does not cover 0
#>
#> Control Group: Never Treated, Anticipation Periods: 0
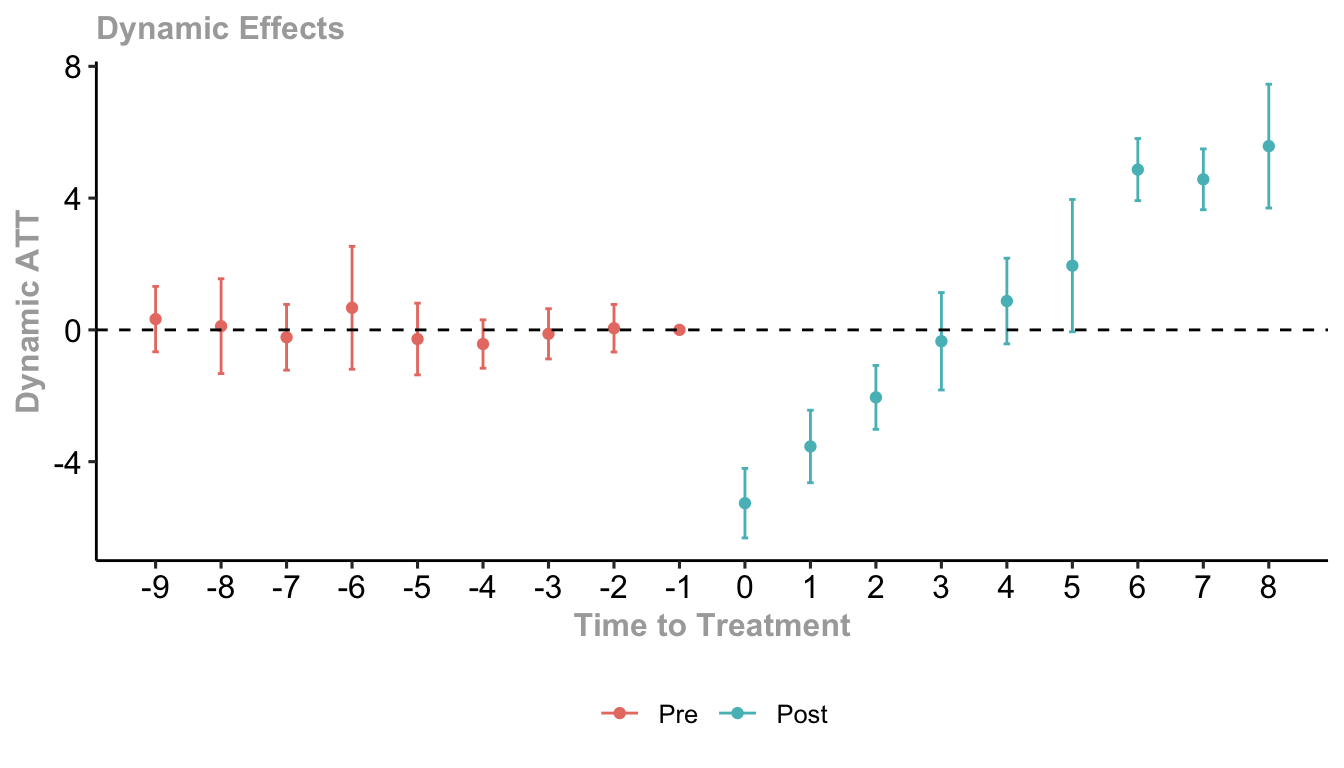
#> Estimation Method: Doubly RobustWe can estimate dynamic treatment effects with the aggte() function and plot with the ggdid() function.
mod |>
aggte(type = "dynamic", na.rm = T) |>
ggdid(
xlab = "Time to Treatment", # x-axis label
ylab = "Dynamic ATT", # y-axis label
title = "Dynamic Effects" # you can include a title string if you want
)
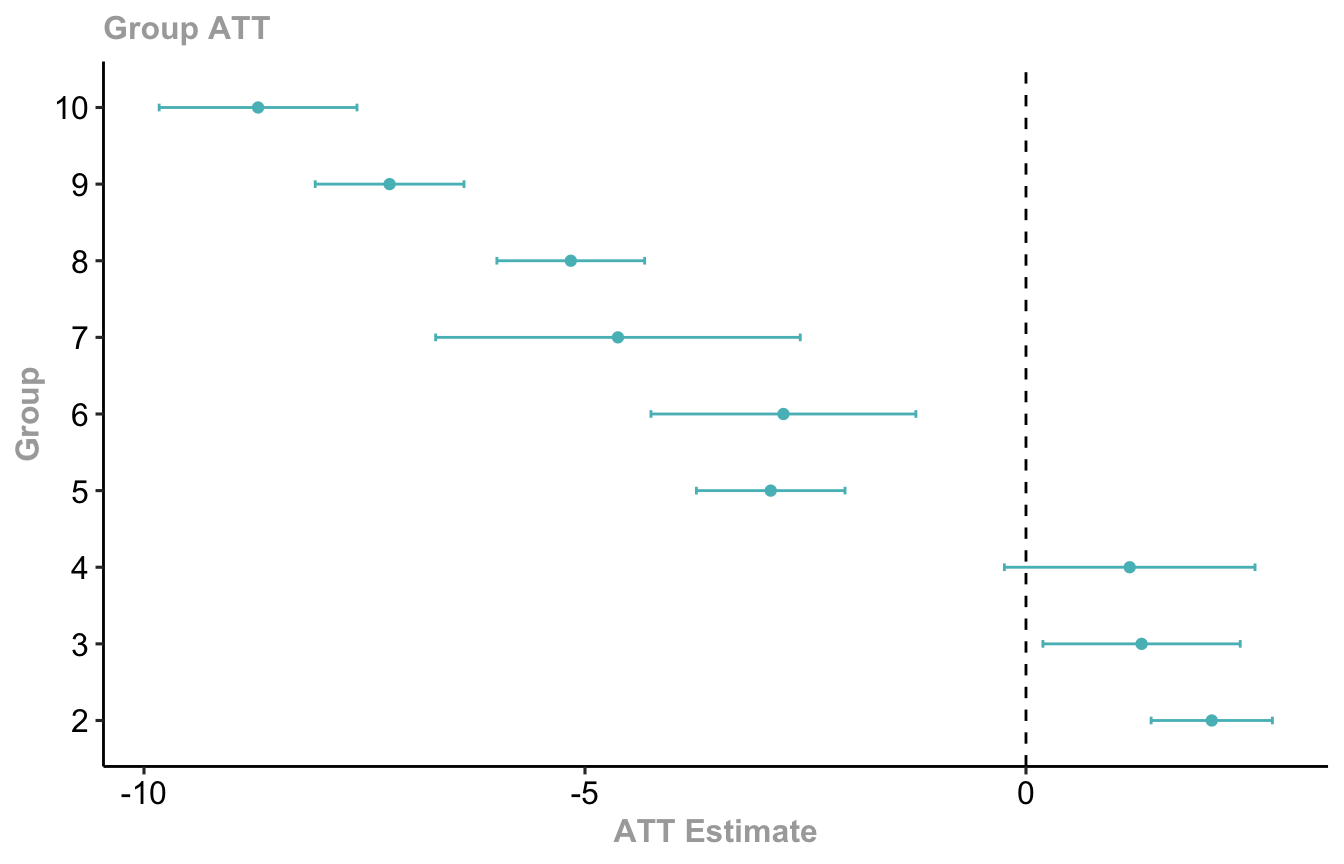
We can also aggregate effects by initial treatment period group, and with the ggdid() function:
mod |>
aggte(type = "group", na.rm = T) |>
ggdid(
xlab = "ATT Estimate", # x-axis label
ylab = "Group", # y-axis label
title = "Group ATT" # you can include a title string if you want
)
Other options for type = include "calendar", which displays the treatment effects grouped by actual (not relative) time period.