install.packages('etwfe')etwfe
R package for extended-twfe (Wooldridge 2021, 2023)
etwfe is a package that implements the extended twfe estimator proposed by Wooldridge (2021, 2023) that solves the bias of the TWFE in staggered settings. Documentation can be found here.
Install the package as follows:
sample code
Start by loading packages and the data:
library(etwfe)
library(readr) # for importing data
df = read_csv('df.csv')We use the etwfe() function to estimate the etwfe model:
mod = etwfe(
fml = outcome ~ covar, # see notes for no covar
tvar = time,
gvar = cohort,
data = df,
vcov = ~ id,
family = NULL # can change to "logit", "negbin", "poisson"
)We can use the emfx() function to aggregate our heterogenous effects into a singular ATT.
mod |> emfx(type = "simple")#>
#> .Dtreat Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> TRUE -1.21 0.148 -8.23 <0.001 52.2 -1.5 -0.925
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSEThe estimate for TRUE is our ATT, and the p-value is given by Pr(>|Z|).
We can use the emfx() function to aggregate dynamic treatment effects. We can then plot these effects with the plot() function. Note that etwfe cannot estimate pre-treatment effects.
mod |>
emfx(type = "event") |>
plot()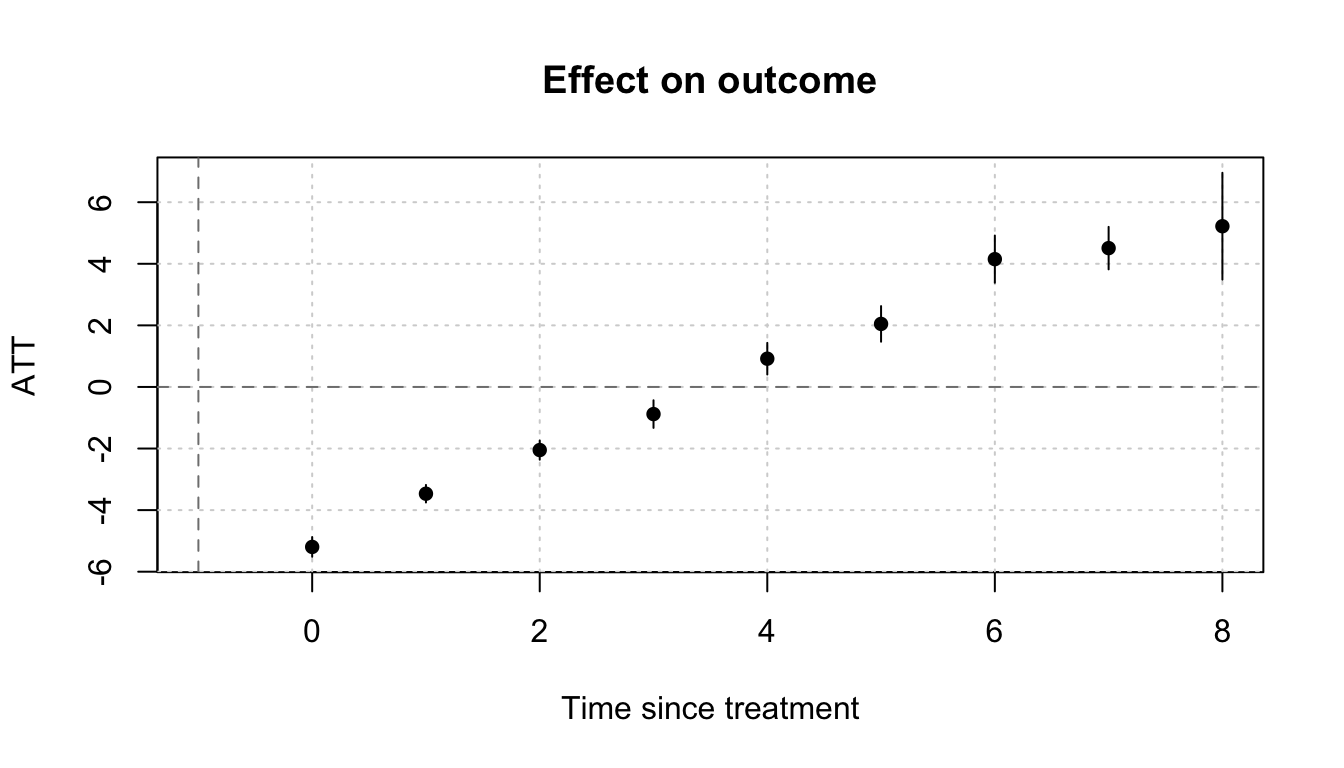
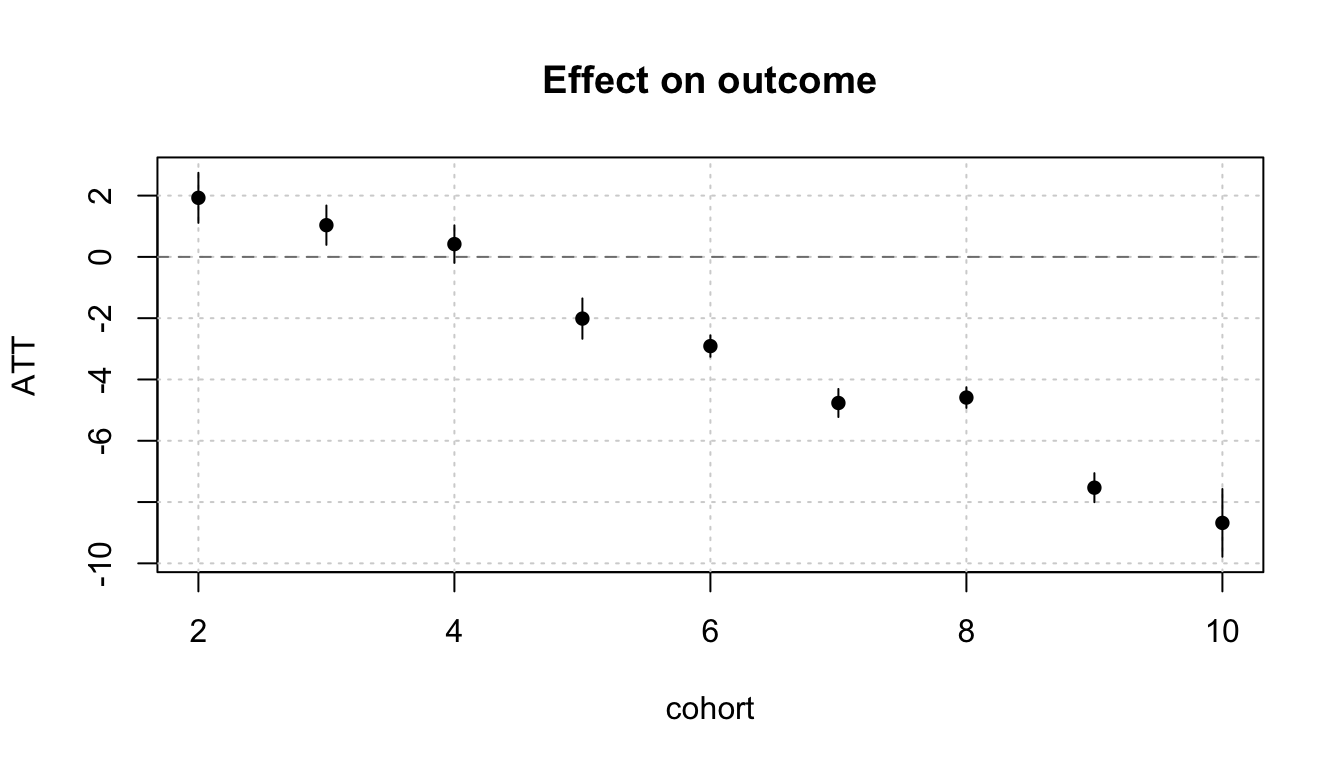
If we are interested in how the ATT differs by year of treatment adoption, we can also aggregate effects by initial treatment period group. We can then plot these effects with the plot() function:
mod |>
emfx(type = "group") |>
plot()