install.packages('DIDmultiplegtDYN')DIDmultiplegtDYN
R package for DIDmultiple estimator (de Chaisemartin and D’Haultfœuille 2024)
DIDmultiplegtDYN is a package that implements the DIDmultiple estimator by de Chaisemartin and D’Haultfœuille (2024), that solves the issues with TWFE, handles non-absorbing and continuous treatments. Documentation can be found here.
Install the package as follows:
sample code
Start by loading packages and the data:
library(DIDmultiplegtDYN)
library(ggplot2) # for plotting
library(readr) # for importing data
df = read_csv('df.csv')We use the did_multiplegt_dyn() function to complete the estimation process.
If you have a continuous or quasi-continuous treatment, or are having issues, set continuous = 1. Covariates for conditional parallel trends are possible to include, but they are used differently, and may cause issues.
mod = did_multiplegt_dyn(
# required arguments
df = df,
outcome = "outcome",
group = "id", # Note: group here refers to unit, not cohort
time = "time",
treatment = "treat",
effects = 4, # Number of post-treatment periods dynamic effects
placebo = 4, # Number of pre-treatment periods of effects
controls = NULL, # optional, vector (string) of covariates.
continuous = NULL, # change to 1 if your treatment is true
graph_off = T
)
mod |> print()#>
#> ----------------------------------------------------------------------
#> Estimation of treatment effects: Event-study effects
#> ----------------------------------------------------------------------
#> Estimate SE LB CI UB CI N Switchers
#> Effect_1 -5.04943 0.45371 -5.93869 -4.16017 675 45
#> Effect_2 -3.25734 0.49253 -4.22269 -2.29199 580 40
#> Effect_3 -2.17826 0.54482 -3.24609 -1.11044 490 35
#> Effect_4 -0.03749 0.50608 -1.02940 0.95442 405 30
#>
#> Test of joint nullity of the effects : p-value = 0.0000
#> ----------------------------------------------------------------------
#> Average cumulative (total) effect per treatment unit
#> ----------------------------------------------------------------------
#> Estimate SE LB CI UB CI N Switchers
#> -2.89921 0.38159 -3.64711 -2.15131 780 150
#> Average number of time periods over which a treatment effect is accumulated: 2.3333
#>
#> ----------------------------------------------------------------------
#> Testing the parallel trends and no anticipation assumptions
#> ----------------------------------------------------------------------
#> Estimate SE LB CI UB CI N Switchers
#> Placebo_1 0.27204 0.49208 -0.69242 1.23651 580 40
#> Placebo_2 -0.72910 0.61928 -1.94287 0.48467 405 30
#> Placebo_3 -0.27729 0.55098 -1.35720 0.80262 250 20
#> Placebo_4 0.00895 1.00786 -1.96642 1.98433 115 10
#>
#> Test of joint nullity of the placebos : p-value = 0.5860
#>
#>
#> The development of this package was funded by the European Union.
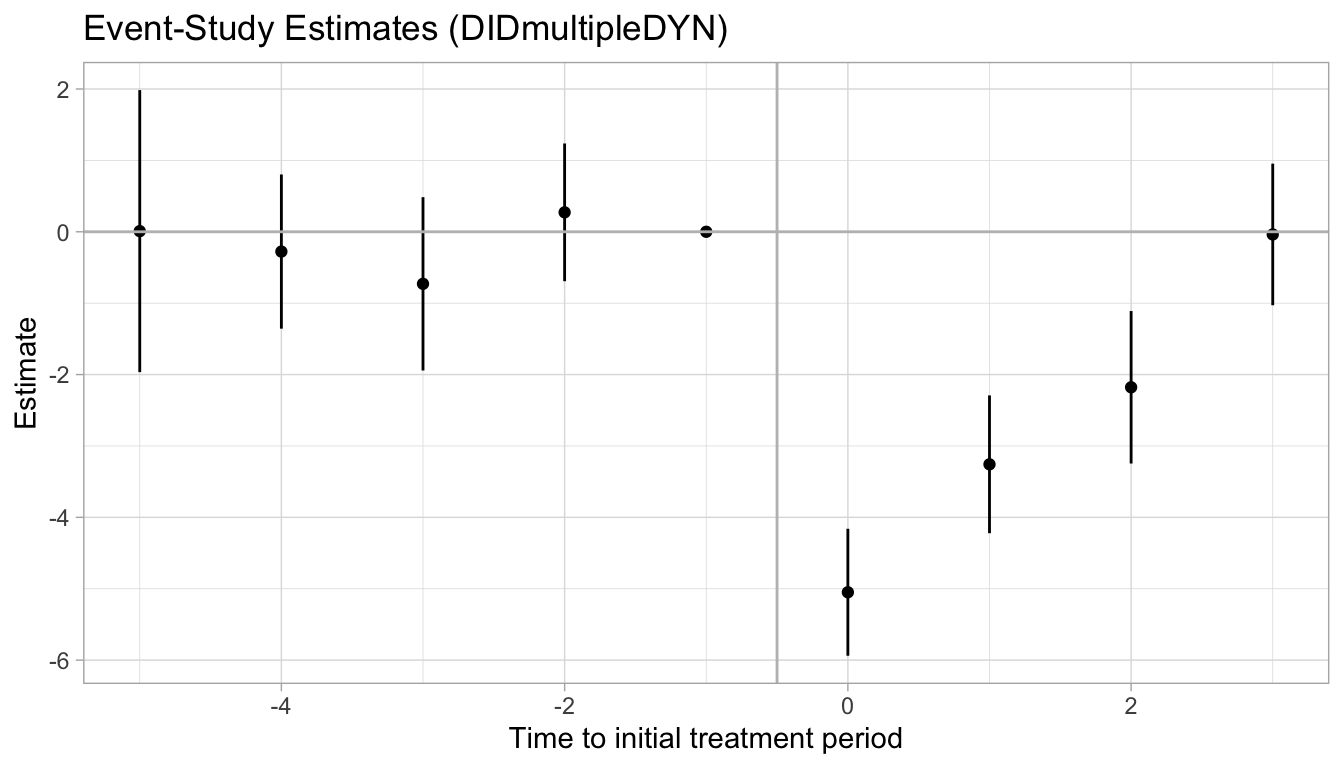
#> ERC REALLYCREDIBLE - GA N. 101043899The Estimation of treatment effects: Event-study effects are the post-treatment dynamic effects. The Average cumulative (total) effect per treatment unit is the ATT estimate. The Testing the parallel trends and no anticipation assumptions is the pre-treatment coefficient estimates.
We can plot the dynamic treatment effects in a plot. The simplest way is to use the default plot - in the original model estimation within did_multiplegt_dyn(), set graph_off = equal to F.
However, the default plot is kind of ugly (in my opinion), and also does not use the conventional numbering of the initial treatment period (it uses t=1 as the initial treatment period). Thus, we can also manually create a ggplot:
# extract effects (the rev() function used because placebo effects are in the opposite order)
effects = c(rev(mod$results$Placebos[,1]), 0, mod$results$Effects[,1]) # extract pre/post effects
lwr = c(rev(mod$results$Placebos[,3]), 0, mod$results$Effects[,3]) # extract lower confint
upr = c(rev(mod$results$Placebos[,4]), 0, mod$results$Effects[,4]) # extract upper confint
# create rel.time variable matching number of pre/post periods
rel.time = c(-5:3)
# create plotting dataframe
plot.df = data.frame(rel.time, effects, lwr, upr)
# ggplot
plot.df |>
ggplot(aes(x = rel.time, y = effects)) +
geom_point() +
geom_linerange(aes(ymin = lwr, ymax = upr)) +
geom_hline(yintercept = 0, color = "gray") +
geom_vline(xintercept = -0.5, color = "gray") +
labs(title = "Event-Study Estimates (DIDmultipleDYN)") +
xlab("Time to initial treatment period") +
ylab("Estimate") +
theme_light()