install.packages('PanelMatch')PanelMatch
R package for PanelMatch DiD estimator (Imai, Kim, and Wang 2023)
PanelMatch is a package that implements the PanelMatch estimator by Imai, Kim, and Wang (2023) that solves the bias of the TWFE estimator in staggered and non-absorbing DiD. Documentation can be found here.
Install the package as follows:
sample code
Start by loading packages and the data:
library(PanelMatch)
library(readr) # for importing data
library(ggplot2) # for plotting
df = read_csv('df.csv')PanelMatch requires us to pre-process the data with the PanelData() function:
Note: id variable should be transformed to an integer variable before starting.
# PanelMatch dislikes tidyverse df's, so do this:
df = df |> as.data.frame()
df.panel = PanelData(
panel.data = df, # your data
unit.id = "id", # your unit var (integer only)
time.id = "time", # your time period var (integer only)
treatment = "treat", # your treatment var
outcome = "outcome" # your outcome var
)Now, we can run the PanelMatch matching process with PanelMatch() to match based on lag-period pre-history:
Lag refers to periods before the treatment in which to match on. Leads refer to periods after the treatment on which to estimate.
match = PanelMatch(
lag = 3, # number of pre-periods to match treat history
panel.data = df.panel, # PanelData generated data
lead = c(0:6), # how many post-treat dynamic effects to estimate
qoi = "att",
refinement.method = "mahalanobis", # set to "none" if no covariates
match.missing = T,
covs.formula = ~ covar, # (optional, can exclude)
placebo.test = T # (optional, but may cause issues)
)To aggregate all the matched comparisons into a singular ATT, we use the PanelEstimate() function.
match |>
PanelEstimate(
panel.data = df.panel, # PanelData object
pooled = T, # tells R to calculate ATT
moderator = NULL # optional. character string for var to calculate heterogenous effects
) |>
print()#> Point estimates:
#> [1] 1.563779
#>
#> Standard errors:
#> [1] 1.189761
#>
#> Estimates produced with 5 observations (non-empty matched sets)The Point estimates are the estimated ATT, and the standard errors are provided.
We can estimate event-study effects with the PanelEstimate() function for post-treatment effects, and the placebo_test() function for pre-treatment effects.
# estimate post-treatment effects
post = match |>
PanelEstimate(
panel.data = df.panel, # PanelData object
pooled = F # tells R to calculate dynamic effects
)
# estimate pre-treatment effects
pre = match |>
placebo_test(
panel.data = df.panel, # PanelData object
lag.in = 3, # should equal lag in PanelMatch()
plot = F
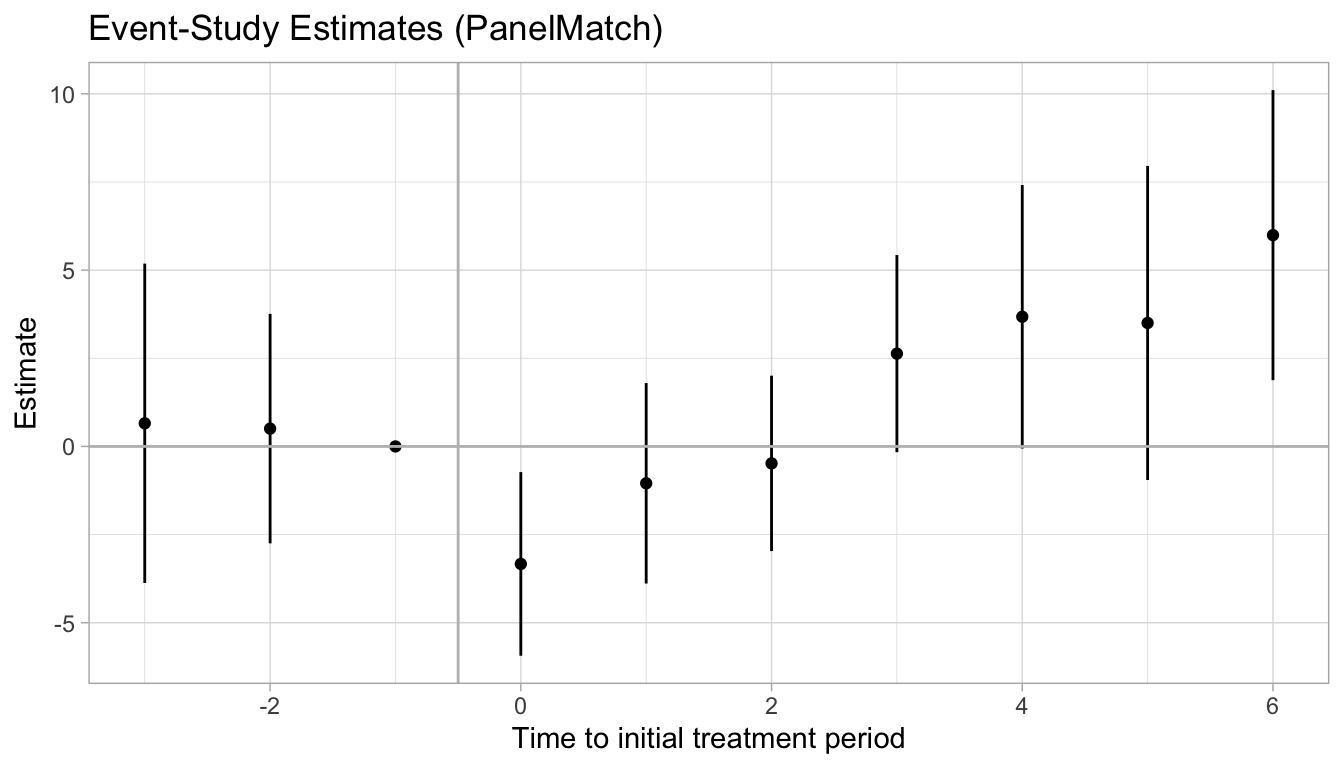
)The built in plotting functions are lackluster, so we will create a manual ggplot to plot the results.
# combine pre and post estimates
effects = c(pre$estimate, 0, post$estimate) # there is no t=-1 effect estimated so add it
se = c(pre$standard.error, 0, post$standard.error)
# create rel.time variable for pre/post periods
rel.time = c(-3:6)
# create results df
results.df = data.frame(rel.time, effects, se)
# first create lwr and upr bounds for se
results.df$se.lwr = results.df$effects - 1.96*results.df$se
results.df$se.upr = results.df$effects + 1.96*results.df$se
results.df |>
ggplot(aes(x = rel.time, y = effects)) +
geom_point() +
geom_linerange(aes(ymin = se.lwr, ymax = se.upr)) +
geom_hline(yintercept = 0, color = "gray") +
geom_vline(xintercept = -0.5, color = "gray") +
labs(title = "Event-Study Estimates (PanelMatch)") +
xlab("Time to initial treatment period") +
ylab("Estimate") +
theme_light()